
The potential products are then automatically analyzed with a BLAST search against user specified databases, to check the specificity to the target intended. The Primer-BLAST tool uses Primer3 to design PCR primers to a sequence template. The deduced amino acid sequence can be saved in various formats and searched against protein databases using BLAST. Sixteen different genetic codes can be used. Open Reading Frame Finder (ORF Finder)Ī graphical analysis tool that finds all open reading frames in a user's sequence or in a sequence already in the database. The MSA Viewer allows users to upload an alignment and set a master sequence, and to explore the data using features such as zooming and changing of coloration.
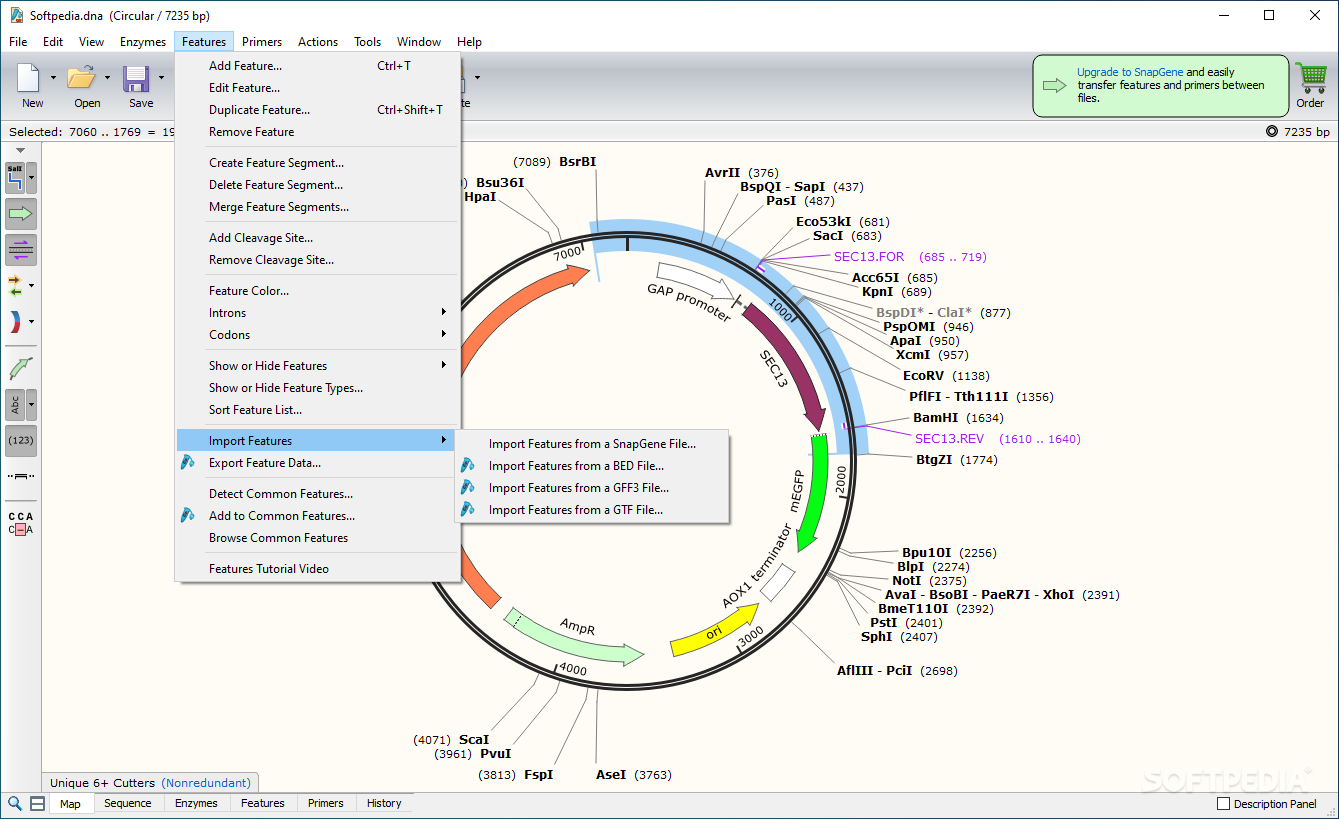
#N GENE SEQUENCE SNAPGENE VIEWER SOFTWARE#
Multiple Sequence Alignment ViewerĪn interactive web application that enables users to visualize multiple alignments created by database search results or other software applications. With Genome Workbench, you can view data in publically available sequence databases at NCBI, and mix these data with your own data. Genome WorkbenchĪn integrated application for viewing and analyzing sequence data.
#N GENE SEQUENCE SNAPGENE VIEWER FULL#
Full results can be downloaded for viewing in NCBI's Genome Workbench graphical viewer, and annotation data for the remapped features, as well as summary data, is also available for download. Options are provided to adjust the stringency of remapping, and summary results are displayed on the web page. NCBI's Remap tool allows users to project annotation data and convert locations of features from one genomic assembly to another or to RefSeqGene sequences through a base by base analysis. This browser also enables the production of displays for publishing. GDV offers easy-to-load analytical track pre-configurations, a menu of data tracks for easy display and customization, and supports upload and analysis of user data. Genome Data Viewer (GDV)Ī genome browser for interactive navigation of eukaryotic RefSeq genome assemblies with comprehensive inspection of gene, expression, variation and other annotations. This tool compares nucleotide or protein sequences to genomic sequence databases and calculates the statistical significance of matches using the Basic Local Alignment Search Tool (BLAST) algorithm. Tool for aligning a query sequence (nucleotide or protein) to GenBank sequences included on microarray or SAGE platforms in the GEO database. CD-Search uses RPS-BLAST (Reverse Position-Specific BLAST) to compare a query sequence against position-specific score matrices that have been prepared from conserved domain alignments present in the Conserved Domain Database (CDD). Identifies the conserved domains present in a protein sequence.
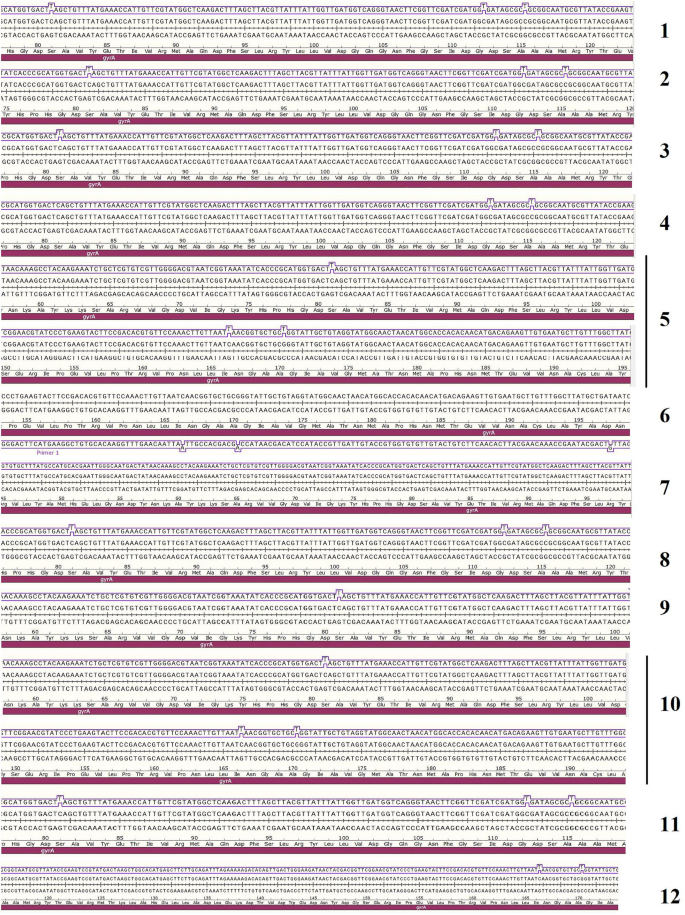
Conserved Domain Search Service (CD Search) COBALTĬOBALT is a protein multiple sequence alignment tool that finds a collection of pairwise constraints derived from conserved domain database, protein motif database, and sequence similarity, using RPS-BLAST, BLASTP, and PHI-BLAST. BLAST can be used to infer functional and evolutionary relationships between sequences as well as to help identify members of gene families. The program compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of matches.
Basic Local Alignment Search Tool (BLAST)įinds regions of local similarity between biological sequences. The default display provides ready navigation to review alignments in the Graphics display. Performs a BLAST search of the genomic sequences in the RefSeqGene/LRG set. Performs a BLAST search for similar sequences from selected complete eukaryotic and prokaryotic genomes.


 0 kommentar(er)
0 kommentar(er)
